Overview
Microbes are often referred to as “the greatest chemists” because they possess a diverse array of metabolic pathways that can produce a wide range natural products and perform complex chemical transformations. For example, the environmental microbes are the primary source of natural products, and the human gut microbiome has a profound impact on human health and well-being. To harness the knowledge and potential of these remarkable chemists, the Li Lab focuses on the developing innovative tools for microbial studies. Our approach combines bioanalytical chemistry, microfluidics, molecular biology, and bioinformatics to construct novel high-throughput single-cell analysis methods, allowing us to gain a deeper understanding of the microbial world.
Our research projects include, 1) single bacteria genomic sequencing, 2) microbial cell-cell interaction, 3) novel biosynthetic gene clusters and metabolites discovery, and 4) microbial-environmental interaction-such as gut microbiome-drug interaction and microbial-plant interaction. The potential outcome of these projects includes the cultivation of “unculturable” species based on acquired knowledge, the identification of interaction-based biocontrol agents, the creation of personalized therapeutics, and new enzymes and chemicals.
Given the multidisciplinary nature of this work, students in our research group are trained in bioanalytical chemistry, microfluidics, single cell genetics, microbiology, medicinal chemistry, and bioinformatics.
Droplet Microfluidics
Droplet microfluidics (left) is a powerful technology that enables the generation and manipulation of millions of nanoliter- to picoliter-sized droplets. It is highly suitable for single-cell analysis, as it allows for the encapsulation of individual cells into nanoliter droplets, creating isolated nano-reaction chambers for precise single-cell analysis. The Li Lab has extensive experience in droplet microfluidics-based single microbial sequencing and high-throughput assays.
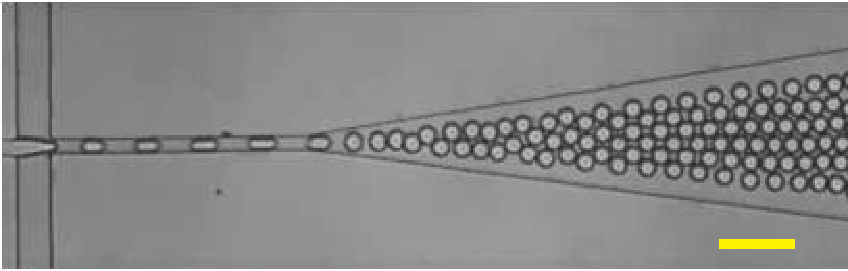
Droplet microfluidics. In a microfluidic channel, droplets are generated when the continuous phase (oil) is injected from the side channel, pinching off the dispersed phase (aqueous) at the junction. Each droplet acts as an independent reaction container for single-cell studies. Droplets can be produced at frequencies exceeding 10 kHz, enabling ultra-high throughput analysis. Scale bar = 100 µm.
Single Microbe sequencing

Single cell sequencing is valuable for dissecting complex systems and uncovering unique features in individual cell types.
However, tools for single microbial cell analysis are limited. We introduce EASi-seq, which adapts a commercially available single cell platform for efficient sequencing of single microbial genomes. EASi-seq enables sequencing of thousands of microbes per run, generating detailed atlases of microbiomes. EASi-seq offers accessible high-quality single cell genomic data for microbiome samples on a commercially available platform.
Cross Domain Taxonomic Analysis
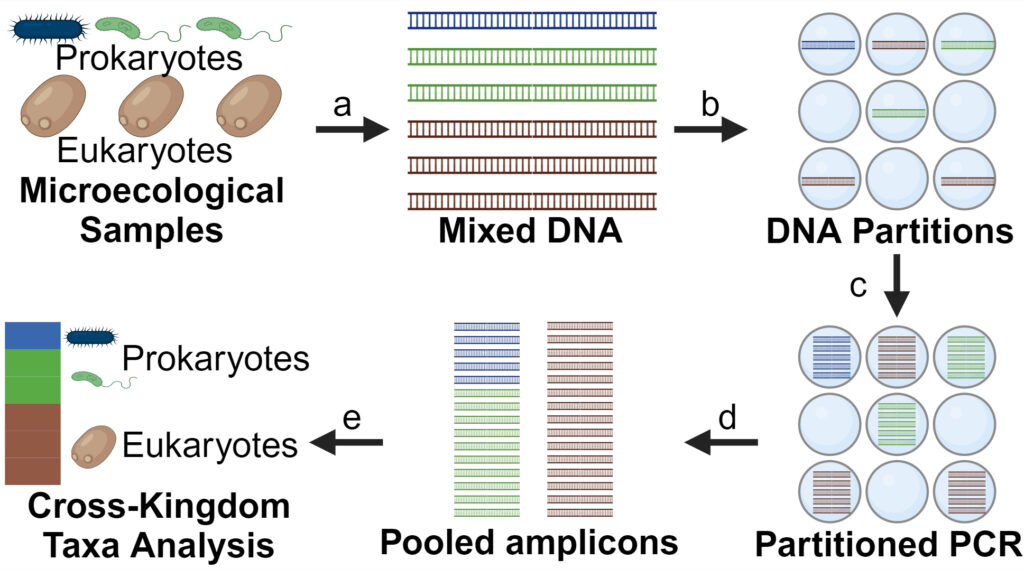
Microbiota are complex communities of prokaryotic and eukaryotic cells, that play distinct roles in maintaining ecosystem balance and influencing host health. A key question and challenge in studying a microbiota is: Which species are present? However, microbial communities are often composed of both culturable and unculturable species, making traditional cultivation methods insufficient. Comprehensive taxonomic analysis, encompassing both prokaryotic and eukaryotic domains, is therefore essential to accurately characterize the diversity and interactions within these microbial ecosystems.
The Li Lab use droplet amplification to addresses the traditional PCR limitations in cross-species classification and developed Partitioned Amplification Multiplexed Amplicon Sequencing (PAM-seq) for cross-domain taxonomic analysis
